réservé à la recherche
Pioglitazone (U-72107) PPARγ Agoniste
N° Cat.S2590
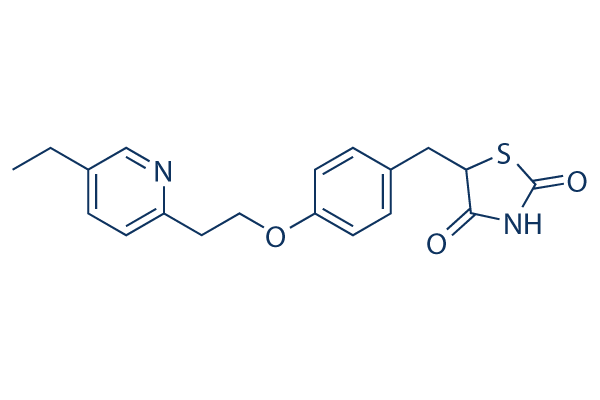
Structure chimique
Poids moléculaire: 356.44
Contrôle qualité
| Cibles apparentées | Dehydrogenase HSP Transferase P450 (e.g. CYP17) PDE phosphatase Vitamin Carbohydrate Metabolism Mitochondrial Metabolism Drug Metabolite |
|---|---|
| Autre PPAR Inhibiteurs | T0070907 GW9662 GW6471 WY-14643 (Pirinixic Acid) GSK3787 GW0742 AZ6102 Harmine Astaxanthin Eupatilin |
Culture cellulaire, traitement et concentration de travail
| Lignées cellulaires | Type dessai | Concentration | Temps dincubation | Formulation | Description de lactivité | PMID |
|---|---|---|---|---|---|---|
| CV-1 | Function assay | In vitro transcriptional activation of Peroxisome proliferator activated receptor gamma (PPAR) expressed in CV-1 cells, EC50=0.69μM | 8576907 | |||
| 3T3-L1 | Function assay | Effective concentration for 50% enhancement of insulin-induced triglyceride accumulation in 3T3-L1 cells, EC50=0.16μM | 9599241 | |||
| CV-1 | Function assay | Activation of peroxisome proliferator activated receptor gamma measured by induction of 50% of maximum alkaline phosphatase activity, transfection assay in CV-1 cells, EC50=0.58884μM | 9836620 | |||
| CV-1 | Function assay | Maximal reporter activity against human Peroxisome proliferator activated receptor gamma Gal4 chimeric in transiently transfected CV-1 cells by functional assay., EC50=0.58μM | 11720854 | |||
| HEK293 | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as induction of receptor interaction with steroid receptor coactivator-1 by EYFP based reporter gene assay | 16680159 | |||
| HEK293 | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as induction of receptor interaction with retinoid X-receptor alpha by EYFP based reporter gene assay | 16680159 | |||
| CV1 | Function assay | Transactivation of PPARgamma in CV1 cells, EC50=0.55μM | 16821769 | |||
| Cos7 | Function assay | 14 hrs | Transactivation of human PPARgamma LBD expressed in african green monkey Cos7 cells co-transfected with fused GAL4-DBD after 14 hrs by Dual-Glo Luciferase reporter gene assay, EC50=0.3μM | 20307981 | ||
| 3T3L1 | Function assay | 100 umol/L | Increase in PPARgamma mRNA levels in mouse 3T3L1 cells at 100 umol/L by RT-PCR | 20392645 | ||
| CHO | Function assay | Activation of Gal4-tagged human PPARgamma expressed in CHO cells by luciferase reporter gene assay, EC50=0.14μM | 20656494 | |||
| HEK293T | Function assay | 1 uM | 24 hrs | Partial agonist activity at human PPARgamma-LBD expressed in HEK293T cells assessed as induction of receptor transactivation at 1 uM after 24 hrs by luciferase reporter gene assay | 21030263 | |
| COS-1 | Function assay | Agonist activity at human PPARgamma ligand binding domain expressed in COS-1 cells co-transfected with Gal4 by luciferase reporter gene assay, EC50=0.39μM | 21130649 | |||
| CHO | Function assay | 24 hrs | Partial agonist activity at human PPARgamma expressed in CHO cells co-transfected with Gal4-responsive luciferase reporter plasmid after 24 hrs by transactivation assay, EC50=0.39μM | 21377875 | ||
| COS7 | Function assay | Modulation of human PPARgamma-LBD expressed in african green monkey COS7 cells co-transfected with Gal4 assessed as activation of transactivation activity by luciferase assay, EC50=0.3μM | 21873070 | |||
| Sf21 | Function assay | Inhibition of human BSEP expressed in plasma membrane vesicles of Sf21 cells assessed as inhibition of ATP-dependent [3H]taurocholate uptake, IC50=0.3μM | 21965623 | |||
| Sf21 | Function assay | Inhibition of Sprague-Dawley rat Bsep expressed in plasma membrane vesicles of Sf21 cells assessed as inhibition of ATP-dependent [3H]taurocholate uptake, IC50=2μM | 21965623 | |||
| HepG2 | Function assay | 20 hrs | Agonist activity at GAL4-tagged human PPARgamma ligand binding domain expressed in HepG2 cells assessed as transactivation after 20 hrs by beta-galactosidase reporter gene assay, EC50=0.57μM | 22341573 | ||
| HEK293 | Function assay | 18 hrs | Transactivation of human GAL4-fused PPARgamma ligand binding domain transfected in HEK293 cells after 18 hrs by dual luciferase reporter gene assay, EC50=0.8μM | 23102891 | ||
| COS7 | Function assay | Transactivation of GAL4-fused human PPARgamma ligand binding domain transfected in african green monkey COS7 cells by luciferase reporter gene assay, EC50=0.2μM | 23130964 | |||
| THP1 | Antiinflammatory assay | 1 hr | Antiinflammatory activity in human THP1 cells assessed as inhibition of PMA-induced MCP-1 secretion preincubated for 1 hr prior PMA-challenge measured after 48 hrs by ELISA, IC50=18.84μM | 23811092 | ||
| THP1 | Antiinflammatory assay | 2 hrs | Antiinflammatory activity in human THP1 cells assessed as inhibition of PMA-induced MCP-1 secretion incubated for 2 hrs prior to PMA challenge measured after 72 hrs by ELISA, IC50=18.6μM | 24531227 | ||
| HEK293 | Function assay | 10 uM | 24 hrs | Transactivation of PPAR-gamma (unknown origin) expressed in HEK293 cells at 10 uM after 24 hrs by luciferase reporter gene assay | 24890090 | |
| HEK293 | Function assay | 18 hrs | Agonist activity at human PPARgamma expressed in HEK293 cells incubated for 18 hrs by luciferase reporter gene assay, EC50=0.21μM | 25333853 | ||
| COS7 | Function assay | Transactivation of human PPARgamma expressed in African green monkey COS7 cells incubated overnight by dual-glo luciferase reporter gene assay, EC50=0.2μM | 26595749 | |||
| THP1 | Function assay | 10 uM | 24 hrs | Upregulation of ABCA1 mRNA expression in human THP1 cells at 10 uM after 24 hrs by qPCR method relative to control | 27220065 | |
| THP1 | Function assay | 10 uM | 24 hrs | Upregulation of ABCG1 mRNA expression in human THP1 cells at 10 uM after 24 hrs by qPCR method relative to control | 27220065 | |
| COS7 | Function assay | 42 hrs | Transactivation of GAL4-fused human PPARgamma ligand binding domain expressed in African green monkey COS7 cells after 42 hrs by dual luciferase reporter gene assay, EC50=0.5μM | 27560282 | ||
| COS7 | Function assay | 42 hrs | Transactivation at Gal4 fused PPARgamma LBD (unknown origin) expressed in African green monkey COS7 cells after 42 hrs by luciferase assay, EC50=0.32μM | 27569195 | ||
| COS7 | Function assay | 42 hrs | Transactivation of Gal4 fused human PPARgamma LBD expressed in African green monkey COS7 cells after 42 hrs by dual luciferase reporter gene assay, EC50=0.38μM | 27918994 | ||
| PC3 | Function assay | 50 uM | 24 hrs | Increase in p21(WAF1) protein expression in human PC3 cells at 50 uM incubated for 24 hrs by Western blot analysis | 28395220 | |
| MDA-MB-231 | Function assay | 50 uM | 24 hrs | Increase in p21(WAF1) protein expression in human MDA-MB-231 cells at 50 uM incubated for 24 hrs by Western blot analysis | 28395220 | |
| HEK293 | Function assay | 24 hrs | Transactivation activity at Gal4 fused full length human PPARgamma LBD expressed in HEK293 cells after 24 hrs by luciferase reporter gene assay, EC50=1.1μM | 28465099 | ||
| HEK293 | Function assay | 4 hrs | Transrepression activity at human PPARgamma expressed in HEK293 cells assessed as inhibition of TNFalpha induced NF-kappaB promoter activity pretreated for 4 hrs followed by TNFalpha stimulation after 3 hrs by luciferase reporter gene assay, IC50=22μM | 28465099 | ||
| SCC15 | Cytotoxicity assay | 50 uM | 24 hrs | Cytotoxicity against human SCC15 cells transfected with PPARalpha siRNA assessed as reduction in cell viability at 50 uM after 24 hrs by resazurin reduction assay | 29031063 | |
| SCC15 | Cytotoxicity assay | 50 uM | 24 hrs | Cytotoxicity against human SCC15 cells transfected with PPARbeta siRNA assessed as reduction in cell viability at 50 uM after 24 hrs by resazurin reduction assay | 29031063 | |
| HepG2 | Function assay | 24 hrs | Agonist activity at PPARgamma in human HepG2 cells assessed as activation of PPRE incubated for 24 hrs by dual luciferase reporter gene assay, EC50=0.24μM | 30001846 | ||
| HEK293BENA | Function assay | 24 hrs | Transactivation of GAL4-fused human PPARgamma transfected in HEK293BENA cells after 24 hrs by steady glo-luciferase reporter gene assay, EC50=2.053μM | 30351933 | ||
| HEK293 | Function assay | 18 hrs | Transactivation of human full length PPARgamma expressed in HEK293 cells after 18 hrs by luciferase reporter gene based luminescence assay, EC50=0.1μM | 30362739 | ||
| 293H | Function assay | 16 hrs | Transactivation of GAL4-DBD fused human PPARgamma ligand binding domain expressed in UAS-bla HEL 293H cells preincubated for 16 hrs followed by FRET substrate addition and measured after 2 hrs by TR-FRET assay, EC50=0.098μM | 30429097 | ||
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in AP1 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in CD36 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in Glut4 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in adiponectin mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| stem cells | Function assay | 5 days | Induction of adipogenesis in human bone marrow-derived mesenchymal stem cells assessed as increase in adiponectin production measured on day 5 in presence of IDX by ELISA, EC50=0.35μM | 30672698 | ||
| HEK293T | Function assay | 18 hrs | Transactivation of full length human PPARgamma2 expressed in HEK293T cells co-expressing PPRE after 18 hrs by luciferase reporter gene assay, EC50=0.25μM | 30676741 | ||
| HEK293T | Function assay | 18 hrs | Transactivation of chimeric Gal4 yeast DBD fused-PPARgamma LBD (unknown origin) expressed in HEK293T cells co-expressing PPRE after 18 hrs by luciferase reporter gene assay, EC50=0.35μM | 30676741 | ||
| HepG2 | Function assay | 30 uM | Binding affinity to NAF1 (unknown origin) expressed in human HepG2 cells assessed as inhibition of mitochondrial respiration at 30 uM | 30770154 | ||
| HepG2 | Function assay | 30 uM | Binding affinity to NAF1 in human HepG2 cells assessed as inhibition of mitochondrial respiration at 30 uM | 30770154 | ||
| COS7 | Function assay | 39 hrs | Transactivation of Gal4-fused human PPARgamma transfected in COS7 cells co-transfected with pGAL5-TK-pGL3 and pRennilla-CMV incubated for 39 hrs by dual luciferase reporter assay, EC50=1.4μM | 31648125 | ||
| K562-IMA[r] | Function assay | 10 uM | 72 hrs | Induction of sensitization to imatinib-induced cytotoxicity in imatinib-resistant human K562-IMA[r] cells assessed as induction of cell death at 10 uM after 72 hrs in presence of 1 uM imatinib by propidium iodide/Triton-X100 dye based FACS analysis | 31648125 | |
| Cliquez pour voir plus de données expérimentales sur les lignées cellulaires | ||||||
Informations chimiques, stockage et stabilité
| Poids moléculaire | 356.44 | Formule | C19H20N2O3S |
Stockage (À partir de la date de réception) | |
|---|---|---|---|---|---|
| N° CAS | 111025-46-8 | Télécharger le SDF | Stockage des solutions mères |
|
|
| Synonymes | AD-4833, U 72107 | Smiles | CCC1=CN=C(C=C1)CCOC2=CC=C(C=C2)CC3C(=O)NC(=O)S3 | ||
Solubilité
|
In vitro |
DMSO
: 20 mg/mL
(56.11 mM)
Water : Insoluble Ethanol : Insoluble |
Calculateur de molarité
|
In vivo |
|||||
Calculateur de formulation in vivo (Solution claire)
Étape 1 : Entrez les informations ci-dessous (Recommandé : Un animal supplémentaire pour tenir compte des pertes pendant lexpérience)
Étape 2 : Entrez la formulation in vivo (Ceci nest que le calculateur, pas la formulation. Veuillez nous contacter dabord sil ny a pas de formulation in vivo dans la section Solubilité.)
Résultats du calcul :
Concentration de travail : mg/ml;
Méthode de préparation du liquide maître DMSO : mg médicament prédissous dans μL DMSO ( Concentration du liquide maître mg/mL, Veuillez nous contacter dabord si la concentration dépasse la solubilité du DMSO du lot de médicament. )
Méthode de préparation de la formulation in vivo : Prendre μL DMSO liquide maître, ajouter ensuiteμL PEG300, mélanger et clarifier, ajouter ensuiteμL Tween 80, mélanger et clarifier, ajouter ensuite μL ddH2O, mélanger et clarifier.
Méthode de préparation de la formulation in vivo : Prendre μL DMSO liquide maître, ajouter ensuite μL Huile de maïs, mélanger et clarifier.
Remarque : 1. Assurez-vous que le liquide est clair avant dajouter le solvant suivant.
2. Assurez-vous dajouter le(s) solvant(s) dans lordre. Vous devez vous assurer que la solution obtenue lors de lajout précédent est une solution claire avant de procéder à lajout du solvant suivant. Des méthodes physiques telles que le vortex, les ultrasons ou le bain-marie peuvent être utilisées pour faciliter la dissolution.
Mécanisme daction
| Targets/IC50/Ki |
PPARγ
|
|---|---|
| In vitro |
La Pioglitazone est métabolisée principalement par le CYP2C8 et, dans une moindre mesure, par le CYP3A4 in vitro. |
| In vivo |
La Pioglitazone atténue significativement la dilatation et le dysfonctionnement de la cavité ventriculaire gauche (VG) par échocardiographie ainsi que la pression télédiastolique VG chez les souris atteintes d'un infarctus du myocarde antérieur étendu. Ce composé normalise partiellement le dP/dt(max) et le dP/dt(min) VG, indices de la fonction contractile VG, qui sont significativement réduits chez les souris atteintes d'IM. Ce composé entraîne une activation réduite de la microglie, une induction réduite des cellules iNOS-positives et moins de cellules positives pour la protéine acide fibrillaire gliale dans le striatum et la substantia nigra pars compacta du modèle murin MPTP de la maladie de Parkinson. Il bloque presque complètement la coloration des neurones TH-positifs pour la nitrotyrosine, un marqueur des dommages cellulaires médiés par le NO. Ce produit chimique (environ 20 mg/kg/jour) atténue l'activation gliale induite par le MPTP et prévient la perte de cellules dopaminergiques dans la substantia nigra pars compacta (SNpc) dans le modèle murin MPTP de la maladie de Parkinson. Il entraîne une réduction du nombre de microglies activées et d'astrocytes réactifs dans l'hippocampe et le cortex de souris transgéniques APPV717I âgées de 10 mois. Le traitement avec ce composé réduit l'expression des enzymes pro-inflammatoires cyclooxygénase 2 (COX2) et oxyde nitrique synthase inductible (iNOS). Il diminue les niveaux d'ARNm et de protéines de la bêta-sécrétase-1 (BACE1), ainsi qu'une réduction de 27% des niveaux du peptide Abeta1-42 soluble. |
Références |
|
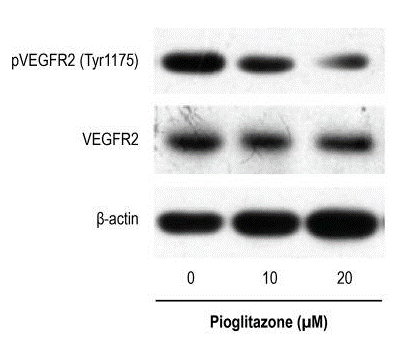
Applications
| Méthodes | Biomarqueurs | Images | PMID |
|---|---|---|---|
| Western blot | p-VEGFR2 / VEGFR Phopsho-RB / RB |
|
29972411 |
Informations sur lessai clinique
(données de https://clinicaltrials.gov, mis à jour le 2024-05-22)
| Numéro NCT | Recrutement | Conditions | Sponsor/Collaborateurs | Date de début | Phases |
|---|---|---|---|---|---|
| NCT05946564 | Not yet recruiting | ANCA Associated Vasculitis|Rapidly Progressive Glomerulonephritis|Crescentic Glomerulonephritis |
Assistance Publique - Hôpitaux de Paris|Ministry of Health France |
July 2023 | Phase 3 |
| NCT05305287 | Recruiting | Non-Alcoholic Fatty Liver Disease|Type 2 Diabetes|Mitochondrial Metabolism Disorders |
The University of Texas Health Science Center at San Antonio|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) |
November 1 2022 | Phase 4 |
| NCT05411965 | Completed | Diabetes Mellitus Type 2 |
Boryung Pharmaceutical Co. Ltd |
April 28 2022 | Phase 1 |
| NCT04501406 | Recruiting | Type 2 Diabetes Mellitus (T2DM)|Nonalcoholic Steatohepatitis |
University of Florida|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) |
December 15 2020 | Phase 2 |
| NCT04535700 | Completed | Type 2 Diabetes |
Fundacion para la Investigacion Biomedica del Hospital Universitario Ramon y Cajal |
September 18 2020 | Phase 4 |
| NCT00904046 | Recruiting | Uric Acid Kidney Stone Disease |
University of Texas Southwestern Medical Center|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)|Takeda Pharmaceuticals North America Inc. |
September 5 2019 | Not Applicable |
Support technique
Tel: +1-832-582-8158 Ext:3
Si vous avez dautres questions, veuillez laisser un message.
